Genetic Regulation of the Human Plasma Proteome in the UK Biobank
- Proteomics
- Read time: 9 minutes
In a landmark study, 13 pharmaceutical companies in conjunction with researchers from the UK Biobank (UKB) present the largest open-access resource of proteomic data analyzing 1,463 proteins across 54,306 UK Biobank participants. A full 10,248 primary genetic associations were identified, of which 85% were newly discovered. This public scientific resource of unprecedented breadth and depth will uncover biological mechanisms underlying genetic discovery and test hypotheses crucial to the development of improved diagnostics and therapeutics for human disease.
The SCALLOP proof of principle: The promise of proteogenomics
In late 2020 a group led by Dr. Anders Mälarstig published a meta-analysis from the SCALLOP (Systematic and Combined AnaLysis of Olink Proteins) consortium1. Currently SCALLOP is comprised of 35 principal investigators from 28 research institutions, with genomic and plasma protein data from over 70,000 patients and healthy controls.
A milestone paper from SCALLOP was published in Nature Metabolism2 and addressed a subset of proteins and samples. Folkersen et al. 2020 is titled “Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals”. Measuring only 90 proteins, over 450 chromosomal locations were identified where the genetic data associated or correlated with the level of a protein measured in circulation. These genomic locations, called protein Quantitative Trait Loci or pQTLs, are what connects genetic variation to level of proteins in the circulation.
The researchers then overlaid 38 diseases and traits that were known to affect a subset of patients among the 30,931 samples tested, and used a statistical technique called Mendelian Randomization to determine which protein (and the genetic region correlated with that protein) has strong evidence of causing a disease.
Of the 451 pQTLs, 25 were identified with high evidence of causality for a given condition; 14 of the 25 confirmed what was known already (that is, already had evidence to be causative for a disease). The remaining 11 of the 25 were novel.
Of the novel findings, proteins were identified to show promise in treating conditions such as rheumatoid arthritis, osteoporosis, prostate cancer and diabetes. These new findings hold enormous potential; first of all to demonstrate a systematic approach to better identify potential drug targets but to also simultaneously gain understanding around the biological mechanism underlying a disease or condition. As a molecular biomarker, the particular identified protein could be measured as a potential diagnostic test, or to monitor disease progression. Therapies can be targeted to modulate the function or level of the protein in the body, and tested for patient benefit in clinical trials. Pivotal to meeting this enormous potential is that the technology used is highly specific to measure the protein expected.
Importantly, in addition to recapitulation of known disease-protein targets, there is the potential for repurposing an existing drug for a new disease. In Folkersen et al., they identified the IL-6 receptor gene (IL-6RA) as not only being highly likely be causal in rheumatoid arthritis, but also the skin condition eczema. The existing on-market drug that targets IL-6 receptor function, a monoclonal antibody called tocilizumab, is currently in use to treat rheumatoid arthritis, and the manufacturer of the drug knows its safety profile.
Can tocilizumab be used to treat eczema? The drug manufacturer now has solid evidence based upon these data to pursue a second indication for this therapeutic to start a new clinical trial for eczema in addition to arthritis.
This SCALLOP study used ‘only’ 90 proteins that were in common among the 30,391 individuals in the consortium. In the preprint by Sun and Whelan et al., 1,463 proteins are measured across 54,306 samples, and several findings are used as illustrations of its power.
The performance of the Olink Explore 1536 assay across 54,306 samples
The UK Biobank announced their use of the Olink Explore 1536 assay in December of 20203, with an original 10 pharmaceutical sponsoring partners. Since then, the number of sponsors has grown to 13 and the total number of samples now numbers over 57,000. In addition, doubling the set of proteins interrogated (to the level measured by the Olink Explore 3072 product) was also announced.
In this preprint, 1,463 proteins identified 10,248 primary genetic associations across 2,928 independent genetic regions. Of these primary pQTLs, 9,098 (85%) have not been identified previously.
Interestingly, 1,162 of the 1,463 proteins (82%) have a cis association (where cis is defined by 1Mb from the gene encoding the protein). The coefficient of variability (CV) of samples run in duplicate had a median of 6.3% suggesting high precision of these measurements over the project. Aside from the many potential biological insights that may be gained from this data, this also provides valuable corroboration of the methodology being applied, since cis-pQTLs provide strong genetic validation that the correct protein is being targeted by the protein detection technology being used.
The breadth of trans-pQTL discoveries do not plateau at more than 50,000 individuals
Through measuring 1,463 proteins, the discovery of new cis-pQTLs plateaus to the number of proteins at approximately 10,000 samples tested (Figure 1, from the paper’s Figure 2E). A total of 1,163 of the 10,248 (11.3%) of the primary genetic associations were in cis, and as you can see from the figure saturation of new cis associations occurs around the 10,000 sample mark.
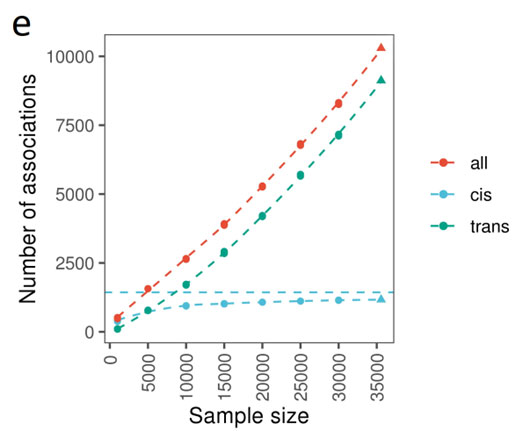
Figure 1: From Figure 2e of Folkersen et al. 2020, number of primary associations against sample size.
In contrast, the number of primary trans-pQTLs continue to be discovered linearly with the number of samples interrogated. The authors emphasize the importance of layering in additional sample types, in particular those from under-represented ancestral populations. The UK Biobank samples from various locations in the United Kingdom, are primarily of Northern European ancestry.
This is a remarkable finding, of fascinating biological implication of the complexity of the genome and how proteins interact genome-wide.
Sensitivity analysis of pQTLs, co-localization with expression QTLs and cis-pQTLs for protein level / RNA level combinations
There may be concerns that individual characteristics such as blood cell composition and BMI may impact the discoverability and reliability of the association between the genetic loci and the protein level. Other aspects of environmental variability such as time of year sampling was taken (seasonal effects) and fasting time before blood collection could affect results. Their analyses determined a “limited impact on the majority of pQTLs”.
When combined with expression quantitative trait loci (eQTL) estimates from the eQTLGen and GTEx (v8) databases of expression levels, only 36% (507 out of 1,425 available genes) shared a causal variant with the RNA transcript expression levels for the gene.
In whole blood, only 11% (111 out of 1,023 available genes) colocalized with an expression QTL. It should be noted that the samples compared across the eQTLGen (RNA levels from blood) and GTEx (RNA levels from tissues) are from different sample populations.
When comparing the lead cis-pQTL for protein level and comparing to the RNA expression level of that same gene, 93.7% of eQTLGen and 83.6% of GTEx protein-expression combinations shared the same direction. The authors note the following: “Pervasive discordant directions of effect for molecular QTLs on gene expression and protein levels are an established phenomenon throughout the human genome”, which have been attributed to various factors such as different rates of protein degradation and protein synthesis on top of different rates of RNA synthesis and degradation.
The example of PCSK9’s effect on lipid levels, cardiovascular and cerebrovascular disease
Various examples are shown in the preprint of proteomic insights into COVID-19 associated loci, a pleiotropic association between the ABO blood group and the fucosyltransferase 2 (FUT2) loci on chromosomes 9 and 19, and connections across the inflammasome pathway.
The authors explore the multiple cis pQTLs as a proxy for the effect of PCSK9 levels, where they demonstrate significant causal effects of increased PCSK9 on increased LDL cholesterol, total cholesterol, and increased risk of coronary heart disease. These findings have been reported previously.
Importantly, effects of increased PCSK9 have also shown to cause decreased HDL cholesterol level, and a significant causal association with increased risk of large artery ischaemic stroke subtype. These findings “have not been substantiated by previous MR studies, likely due to decreased power. These findings extend the corroborated effects observed across multiple randomized clinical trials of PCSK9 inhibitors.”
A new era of proteogenomics has begun
As mentioned above, one of the first proof-of-concept studies in Folkersen et al. 2020 examined 90 proteins across 30,931 individuals demonstrating the potential for understanding novel disease mechanism, drug discovery, and drug repurposing. In this pre-print, examining 1,463 proteins across 54,306 individuals is a landmark to true population proteogenomics.
Hailed as a “spectacular resource” with “unprecedented breadth and depth”, a new age of proteogenomics has begun.
Reference Links:
- The SCALLOP Consortium page. https://www.olink.com/our-community/scallop/
- Folkersen et al. 2020 Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab 22, 1135-1148 https://doi.org/10.5281/zenodo.2615265
- “Olink to provide proteomics data from 53,000 UK Biobank participants” https://www.olink.com/news/olink-to-provide-proteomics-data-from-53000-uk-biobank-participants/
- Sun BB and Whelan CD et al. bioRxiv 2022.06.17.496443 (2022) Genetic regulation of the human plasma proteome in 54,306 UK Biobank participants doi: https://doi.org/10.1101/2022.06.17.496443
How the proteome behaves in healthy individuals
Clinical research, Multiomics
To achieve the goal of precision medicine, not only do different molecular profiles need to be understood in disease populations, but they must also be understood in the context of healthy populations.
Key proteomics publications from 2020
Proteomics
Welcome to the first post of the all-new weekly Olink to Science! Our customer survey revealed that you would like to know more about the many publications, research, and other science happening at Olink, therefore this blog aims to do just that: keep you informed on the exciting science taking place with our technology.
Protein biomarkers are crucial in early detection of cancer
Clinical research, Oncology, Protein biomarkers
A central premise of precision medicine is to identify biomarkers indicative of disease transitions early on. This is especially important in cancer where early treatment intervention could increase a patient’s chance of survival and reduce the probability of cancer recurrence.
Using PEA and RNA-Seq to study disease pathology
Clinical research, Proteomics
The following study illustrates how transcriptomics and proteomics complement one another to clarify the pathology of a complex, and little understood disease. Atopic dermatitis (AD) is the most common chronic skin condition affecting up to 20% of children and 7-10% of adults, depending on the population.
Olink protein biomarker panel indicates fermented foods fight inflammation
Inflammation, Proteomics
Could food be used to fight chronic disease?
Study identifies proteins involved in immunotherapy response
Oncology, Proteomics
'Ultimately, it is all about understanding and treating patients better in the future.'
Proteins diagnostic of lung cancer up to 5 years before disease onset
Oncology
An earlier Olink to Science blog post covered some amazing research that found that certain blood protein biomarkers have the potential to predict cancer up to 3 years before diagnosis. This may also be the case for lung cancer, as detailed in a recent study by Dagnino and her colleagues, where elevated levels of CDCP1 were detected in participants of a cohort who later developed the disease.
Utilizing proteogenomics technology for novel drug target discovery
Drug discovery & development
High-throughput multiplexed proteomic technology is leading the way to the latest developments in pre-clinical disease analysis in drug discovery. The pharmaceutical industry is now increasing its efforts in the discovery of novel drug targets by using protein quantitative trait loci (pQTLs), which allows for a more confident inference of disease causality and associated protein regulation.
Developing a high-performance biomarker panel for Alzheimer’s disease
Clinical research, Neurology, Protein biomarkers
A simple search of the term ‘scourge of Alzheimer’s Disease’ brings up over half a million website hits. A major disease, about 15% of us that reach the age of 67 to 74, and 44% of those 75 to 84 will develop AD.
How proteomics helped diabetic kidney disease research advance
Clinical research, Proteomics
Dr. Krolewski and his team at the Harvard Medical School found 56 proteins to be significant in diabetic kidney disease patients. Potentially, these could serve as prognostic biomarkers for disease progression and treatment response. This is how adding proteomics to the methodologies elevated their research.









