Biological insights revealed by integrating genomics and proteomics data
- Drug discovery & development , pQTLs , Proteogenomics , Proteomics
- Read time: 8 minutes
New approaches to investigate proteomics at population scale are empowering genomics research. In this blog post, learn how novel drug targets and new insight into disease pathways are being revealed by integrating genomics and proteomics data.
Amid a rich history of Genome-Wide Association Studies (GWAS) and a wealth of genetic information, new approaches to investigate proteomics at population scale are empowering researchers to identify proteins and pathways that are likely to be causal in disease.1 One such effort is the UK Biobank, which began in 2006 as a long-term, large-scale biomedical research resource and database collecting in-depth genetic and health information from half a million participants. In 2020, the UK Biobank Pharma Proteomics Project (UKB-PPP) was formed, with the intention to measure proteins in tens of thousands of plasma samples. The project used the Olink® Explore 1536 platform to measure ~1500 proteins.
In a 2022 pre-print, the UKB-PPP identified over 8,000 novel proteogenomic associations between genetic variants and circulating protein levels in 54,306 participants.2 They also identified over 2,000 previously reported proteogenomic associations.
These proteogenomic associations are also known as protein quantitative loci or pQTLs, where a cis-pQTL refers to the association between a variant proximal to the gene encoding the protein being measured. Cis-pQTLs were identified for 85% of the proteins evaluated in the study.2 Considered a surrogate measure of specificity, cis-pQTLs provide strong genetic evidence that the right protein is being measured and highlights the exceptional specificity of Olink® technology.
A trans-pQTL refers to the regulation of a protein-of-interest by a genetic variant that is distal to the gene encoding the protein. Trans-pQTLs are of great potential importance to identify novel regulatory pathways linked to proteins associated with specific phenotypes. Interestingly, the number of trans-pQTLs continued to increase with increasing sample size (Figure 1). 2 The UKB-PPP is now measuring an additional ~1500 plasma proteins with the expanded Explore library (i.e., Olink® Explore 3072).
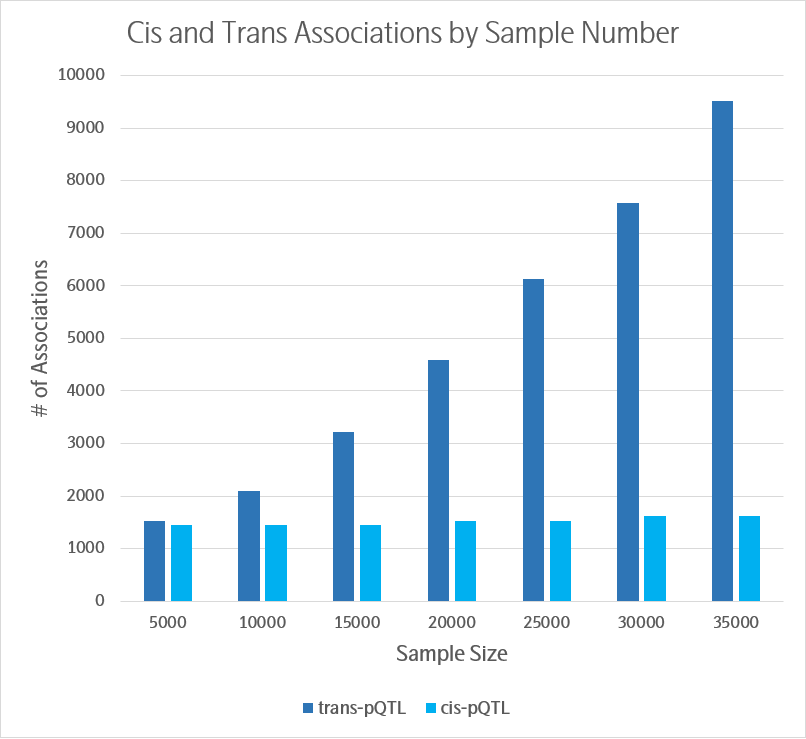
Figure 1. Number of primary cis- and trans-pQTL associations as a function of sample size. Adapted from reference 2, Figure 2e.
Integration of genomics with proteomics was also performed by Koprulu et al. at the University of Cambridge.3 Nearly 3000 serological proteins were measured with Olink Explore 3072, with most of the cis-pQTLs (96.9%) identified in Cohort #1 (n = 1,180) validated in Cohort #2 (n = 1707). Moreover, 256 of the cis-pQTLs were novel. The study also demonstrated the genetic regulation of 575 specific health outcomes via 224 cis-pQTLs. It is noteworthy that many of the cis-pQTLs identified with Olink Explore were not detected in earlier studies with other proteomic technologies, even when those studies had significantly larger sample sets.
Data from the UKB-PPP and the study by Koprulu et al. offer opportunities to accelerate the development of more effective therapeutics and elucidate biological mechanisms that underlie disease. Indeed, the SCALLOP (Systematic and Combined AnaLysis of Olink Proteins) Consortium has collected genetic and proteomic data and clinical phenotypes from 70,000 patients representing 45 cohort studies.4 Seminal work by the SCALLOP Consortium, which includes 35 investigators from 28 institutions, revealed 451 primary genetic associations, discovered 25 causal proteins representing novel drug targets, and suggested 18 drug repurposing opportunities.1 These 25 links to causal proteins bridge the gap between genetic associations and tangible disease pathology. This milestone paper from SCALLOP provides confidence in a systematic approach to therapeutic target discovery by integrating genomics and proteomics data.
To learn more, download the white paper, “Empower genomics with proteomics,” using the button below, or contact us today at info@olink.com.
References
- Folkersen L and Malarstig A et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nature Metab 2020 2(10):1135-1148. doi:10.1038/s42255-020-00287-2
- Sun BB and Whelan CD et al. Genetic regulation of the human plasma proteome in 54,306 UK Biobank participants. bioRxiv 18 June 2022 doi:10.1101/2022.06.17.496443
- Koprulu M, Carrasco-Zanini J, Wheeler E, et al. Proteogenomic links to human metabolic diseases. Nature Metabolism 2023. doi: 10.1038/s42255-023-00753-7
- http://www.scallop-consortium.com/
How the proteome behaves in healthy individuals
Clinical research, Multiomics
To achieve the goal of precision medicine, not only do different molecular profiles need to be understood in disease populations, but they must also be understood in the context of healthy populations.
Key proteomics publications from 2020
Proteomics
Welcome to the first post of the all-new weekly Olink to Science! Our customer survey revealed that you would like to know more about the many publications, research, and other science happening at Olink, therefore this blog aims to do just that: keep you informed on the exciting science taking place with our technology.
Protein biomarkers are crucial in early detection of cancer
Clinical research, Oncology, Protein biomarkers
A central premise of precision medicine is to identify biomarkers indicative of disease transitions early on. This is especially important in cancer where early treatment intervention could increase a patient’s chance of survival and reduce the probability of cancer recurrence.
Using PEA and RNA-Seq to study disease pathology
Clinical research, Proteomics
The following study illustrates how transcriptomics and proteomics complement one another to clarify the pathology of a complex, and little understood disease. Atopic dermatitis (AD) is the most common chronic skin condition affecting up to 20% of children and 7-10% of adults, depending on the population.
Olink protein biomarker panel indicates fermented foods fight inflammation
Inflammation, Proteomics
Could food be used to fight chronic disease?
Study identifies proteins involved in immunotherapy response
Oncology, Proteomics
'Ultimately, it is all about understanding and treating patients better in the future.'
Proteins diagnostic of lung cancer up to 5 years before disease onset
Oncology
An earlier Olink to Science blog post covered some amazing research that found that certain blood protein biomarkers have the potential to predict cancer up to 3 years before diagnosis. This may also be the case for lung cancer, as detailed in a recent study by Dagnino and her colleagues, where elevated levels of CDCP1 were detected in participants of a cohort who later developed the disease.
Utilizing proteogenomics technology for novel drug target discovery
Drug discovery & development
High-throughput multiplexed proteomic technology is leading the way to the latest developments in pre-clinical disease analysis in drug discovery. The pharmaceutical industry is now increasing its efforts in the discovery of novel drug targets by using protein quantitative trait loci (pQTLs), which allows for a more confident inference of disease causality and associated protein regulation.
Developing a high-performance biomarker panel for Alzheimer’s disease
Clinical research, Neurology, Protein biomarkers
A simple search of the term ‘scourge of Alzheimer’s Disease’ brings up over half a million website hits. A major disease, about 15% of us that reach the age of 67 to 74, and 44% of those 75 to 84 will develop AD.
How proteomics helped diabetic kidney disease research advance
Clinical research, Proteomics
Dr. Krolewski and his team at the Harvard Medical School found 56 proteins to be significant in diabetic kidney disease patients. Potentially, these could serve as prognostic biomarkers for disease progression and treatment response. This is how adding proteomics to the methodologies elevated their research.









