Collaborative effort to investigate the plasma proteomic signatures of COVID-19 positive patients
Olink is immensely proud of our contribution to one of the largest longitudinal COVID-19 studies together with the prestigious Massachusetts General Hospital (MGH).
What the investigators say
“We are grateful to have entered this collaborative effort with Olink Proteomics to investigate the plasma proteomic signatures of COVID-19 patients and with Olink’s help have profiled over 1400 plasma proteins in our entire cohort. We hope together we can provide the clinical and scientific community with a rich dataset for further investigation of pathways underlying severe disease that may be the basis for early diagnosis and clinical intervention. As such, we are eager to share our data broadly with the scientific community to augment others’ findings and to accelerate discovery that may lead to new therapies and a better understanding of the underpinnings of COVID-19.”
Michael R. Filbin, M.D., M.S., Department of Emergency Medicine, Massachusetts General Hospital
A paper reporting on the findings from this study is now published in Cell Reports Medicine : Read article
You may also be interested to read an article related to this study that Dr. Filbin contributed to Drug Target Review – read the article
Access data from the study
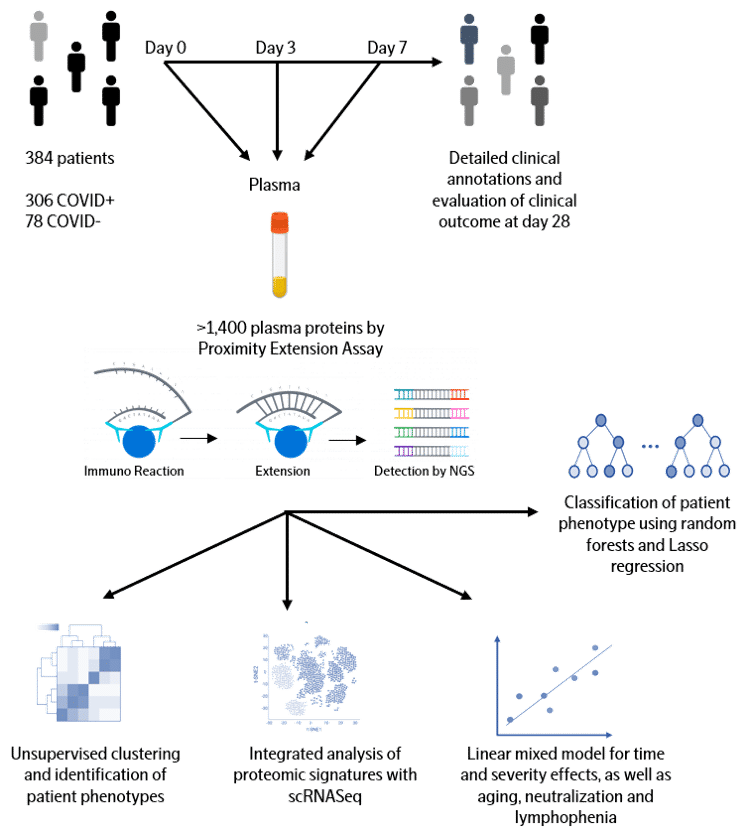
Data from one of the largest longitudinal COVID-19 studies is now publicly available, with data run on the new Olink® Explore 1536 platform. The samples are from a study which included patients 18 years or older with a clinical concern for COVID-19 upon Emergency Department arrival, and who had acute respiratory distress. Of the 384 patients enrolled, 306 tested COVID-19 positive and the 78 COVID-negative patients were included as a comparator group. In collaboration with the group from the MGH, the raw data from the study (protein measurements and essential clinical parameters) is accessible via the Olink website for everyone in the global scientific community. Please use the button below to access the data.
Citing this data
To acknowledge the source of this data in publications, presentations or reports etc., please use the following formulation:
”Data provided by the MGH Emergency Department COVID-19 Cohort (Filbin, Goldberg, Hacohen) with Olink Proteomics”.
Background
COVID-19 patients exhibit a broad range of clinical phenotypes, ranging from asymptomatic to fatal disease. To identify cellular and immune responses associated with outcome, a group of clinicians and immunologists at Massachusetts General Hospital (MGH) enrolled acutely-ill patients in the Emergency Department in a large, urban, academic hospital in Boston (with institutional review board approval) during the COVID-19 surge from late winter to early spring of 2020. The cohort included patients 18 years or older with a clinical concern for COVID-19 upon ED arrival, and with acute respiratory distress with at least one of the following: 1) tachypnea ≥ 22 breaths per minute; 2) oxygen saturation ≤ 92% on room air; 3) a requirement for supplemental oxygen; or 4) positive-pressure ventilation. Of 384 patients enrolled, 306 (80%) tested positive and 78 patients did not test positive for SARS-CoV-2. COVID-19-positive patients had their blood sampled on days 0, 3, and 7, while virus-negative patients had sampling only on day 0 and served as a comparator group. Comprehensive clinical data were collected on this cohort, including 28-day outcomes classified according to the World Health Organization (WHO) COVID-19 outcomes scale.
Initial analysis
Olink Proteomics joined forces with MGH to quantify the abundance of over 1400 proteins in the plasma of each patient from the cohort of 306 COVID-19 patients and 78 virus-negative patients. To accelerate global research in COVID-19 pathogenesis, we are now sharing the entire dataset with the clinical and scientific community. We hope this unique and large data will be integrated with other datasets to validate findings, develop new insights into what drives COVID-19 disease and ultimately lead to biomarkers for better clinical decision-making and new therapeutic targets.
About the Olink technology used in this study
Olink Proximity Extension Assay (PEA) is a technology developed for high-multiplex analysis of proteins using just a few microliters of samples – view technology video
The full library (Olink Explore 1536) is consisting of 1472 proteins and 48 controls assays divided into four 384-plex panels focused on inflammation, oncology, cardiometabolic and neurology proteins. In each of the four 384-plex panels, overlapping assays of IL-6, IL-8 (CXCL8) and TNF are included for QC purposes.
Library content is based on target selection of low-abundant inflammation proteins, actively secreted proteins, organ-specific proteins leaked into circulation, drug targets (established and from ongoing clinical trials) and proteins detected in blood by mass spectrometry.

Want to know more about Olink Explore 1536?
If you have any questions regarding the high-throughput, high-multiplex protein biomarker discovery platform with NGS readout that was used in this study, please contact us using the button below:
Acknowledgements
Olink would like to extend our sincere appreciation and thanks to all the outstanding researchers involved in this project. In particular, we would like to thank our close collaborators from the Massachusetts General Hospital (MGH) and the Broad Institute below.

We would also like to acknowledge the valuable contributions of the entire team:
Collection Team: Kyle Kays, Kendall Lavin-Parsons, Blair Alden Parry, Brendan Lilley, Carl Lodenstein, Brenna McKaig, Nicole Charland, Hargun Khanna, Justin Margolin
Processing Team: Moshe Sade-Feldman, Anna Gonye, Irena Gushterova, Tom Lasalle, Nihaarika Sharma, Brian C. Russo, Maricarmen Rojas-Lopez, Kasidet Manakongtreecheep, Jessica Tantivit, Molly Fisher Thomas
Data analysis: Arnav Mehta, Alexis Schneider
Principal Investigators: Michael R. Filbin, Alexandra-Chloe Villani, Nir Hacohen, Marcia Goldberg
Questions about Olink and COVID-19?
If you have any questions regarding this study, or anything else regarding the use of Olink technology in COVID-19 research, please contact us using the simple form below: