A new era for high-throughput proteomics
Several factors are driving this paradigm shift
• A growing culture of open collaboration and information sharing
• Big-data analysis tools
• Access to very large, well-characterized biobank sample collections
• Advances in proteomics with the ability to provide reliable data at scale
Genomics has not had the transformative effects on therapeutic medicine originally envisaged.
Empower genomics with proteomics - “proteogenomics”

Technical challenges have been overcome by next-generation proteomics

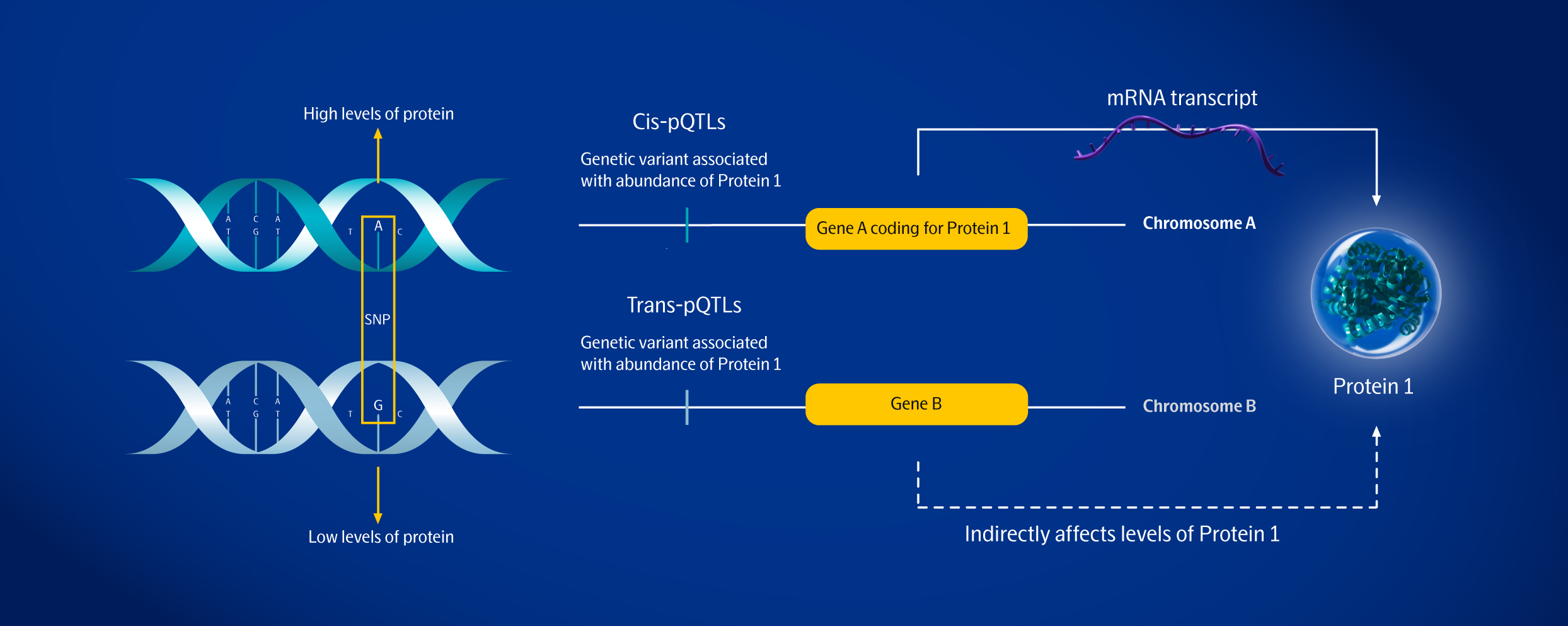
 Identifying robust drug targets with disease causality
Identifying robust drug targets with disease causality
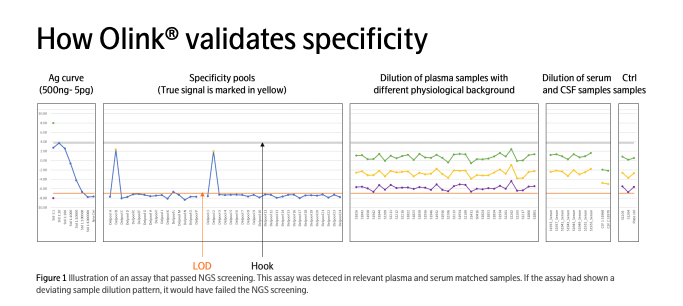
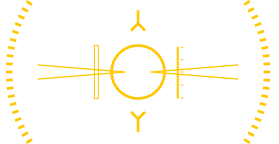 Maintaining specificity in multiplex protein assays
Maintaining specificity in multiplex protein assays
 Scaling up proteogenomics to population-scale studies
Scaling up proteogenomics to population-scale studies
eBook
Empowering genomics with high-throughput proteomics
Take this opportunity to stay at the forefront of this groundbreaking field by reading about the latest developments. See how leading scientists are already implementing high-throughput proteomics to shape the future of precision medicine.
Introducing
Olink® Explore HT
Capture true biological insights with proven specificity. At any scale. Perform high-throughput biomarker discovery with ease to gain an understanding of disease at the protein level.
Accelerate your approach to proteomics with Olink® Insight
An open-access online portal that helps you understand and interpret proteomics data for faster insights – includes Visual Pathway Browser and Disease Atlas, Olink Flex Panel builder and panel selection guide, as well as Publication Explorer and Automatic Annotations.