Mapping Neurological Disease with Proteomics: Advances from the UK Biobank-Pharma Proteomics Project
Protein biomarkers offer tremendous potential to transform the diagnosis, prognosis, and management of neurological disorders such as Alzheimer’s disease, Parkinson’s disease, and multiple sclerosis. However, understanding the intricate mechanisms of neurological diseases requires further studies of the human genome and proteome at the population level. The development of reliable biomarkers has also been hindered by the need for highly sensitive and specific assays. Overcoming these hurdles is essential to advance precision medicine in neurology.
A major step forward came with the pilot phase of the UK Biobank-Pharma Proteomics Project (UKB-PPP), a pioneering initiative that integrated large-scale proteomic profiling using the Olink® Explore 3072 platform with genetic and clinical data from over 50,000 UK Biobank participants. This unique dataset has enabled groundbreaking discoveries at the intersection of genomics and proteomics, significantly deepening our understanding of complex diseases and supporting more personalized medical approaches. The scale and richness of the UK Biobank make it an unparalleled resource for researchers, unlocking insights that were previously unattainable.
Building on the success of the pilot phase, it was announced in January 2025 that Olink® Explore HT (covering over 5,400 proteins) would be used to profile all 500,000 UK Biobank participants. This expanded dataset promises to reveal even more about the intricate connections between proteins and disease, laying the groundwork for targeted therapies and enhanced diagnostic tools.
Among the many therapeutic areas where the UKB-PPP has driven major advances, neurology stands out. With approximately 15,000 neurological disorders represented (Table) and around 50 related studies published to date using UKB-PPP data, the field has already seen significant progress in biomarker discovery for early diagnosis, patient stratification, and disease pathophysiology. In the sections that follow, we will highlight how the UKB-PPP has catalyzed transformative research in neurological science.
Table. Number of cases of different neurological diseases in the UKB-PPP
| Disease / Disorder | Number of cases (UKB-PPP) |
| Depression | 5,446 |
| Pain | 3,477 |
| Sleep disorders | 1,872 |
| Stroke | 893 |
| Epilepsy | 807 |
| Alzheimer’s disease & related dementias | 690 |
| Parkinson’s disease | 342 |
| Motor neuronal disease | 232 |
| Multiple sclerosis | 217 |
| Schizophrenia | 130 |
| Total | ~15,000 |
Prediction of future dementia in healthy individuals
Early prediction and detection of neurodegenerative diseases provides the best opportunity for prevention and treatment. Large-scale population health studies enabled by the UKB-PPP have advanced the prediction of dementia in healthy individuals, up to 10 years before diagnosis (1). Researchers from Shanghai Medical College leveraged their access to UKB-PPP data to perform an impactful in silico investigation to identify predictive risk markers and signatures for all-cause dementia (ACD), Alzheimer’s disease (AD) and vascular dementia (VaD). After identifying 4-11 significant predictive analytes for those disorders, the combination of either GFAP or GDF15 with demographic variables enabled accurate prediction of ACD, AD, and VaD. When considering time to diagnosis from blood draw, combining GFAP with demographic data provided accurate prediction of future dementia over 10 years for ACD and AD. GFAP and NEFL also began to change significantly up to at least 10 years before incident dementia was diagnosed.
Utilizing a data-driven proteomics strategy, we innovatively identified important plasma biomarkers for future dementia prediction from the largest prospective community-based cohort with long-term follow-up to date. These findings are poised to yield significant implications for screening people at high risk for dementia and for early intervention.
Understanding the pathology of Parkinson’s disease with multi-cohort validation
With the aim of identifying predictive biomarkers for Parkinson’s disease (PD), another recent study performed Mendelian randomization to identify predictive and causal proteins linked to PD 2. UKB data indicated 38 proteins associated with incident PD over a follow-up period of 14.5 years. Six of the top 10 most significant proteins were fully replicated in a Parkinson’s Progression Markers Initiative (PPMI) validation cohort. ITGAV, HNMT, and ITGAM showed consistent significant correlations with incident PD across three baseline-to-diagnosis intervals. Moreover, the top 16 associated proteins and demographic factors were able to predict incident PD in subgroup analyses of up to 5 years, and over 5 years, which were further validated in the PPMI cohort.
By characterizing the temporal evolution of sporadic PD, we address key gaps in understanding early-stage pathology, aiding biomarker and therapy development.
The largest plasma proteomic screen of multiple sclerosis – support for known associations and novel biomarkers
Blood-based protein biomarkers are attractive for the diagnosis, monitoring, and prognostication of multiple sclerosis (MS); however, they are not routinely used for the investigation or management of MS. Markers of MS risk and disease severity were evaluated in 407 prevalent MS cases in the UKB that had well characterized clinical data (3). Seventy-two proteins were associated with MS, including several of which identified in previous studies. Among the downregulated proteins, Granzyme A (GZMA) was an entirely novel finding as a MS biomarker. Pathway analysis indicated enrichment of cytokines, cytokine receptors, and lysosomal processing proteins in MS, as well as the MS-associated decrease in proteins involved in leukocyte migration, interaction with the extracellular matrix, regulation of the actin cytoskeleton, and cell–cell adhesion (Figure). The data were also directionally concordant with MS for ~83% of overlapping proteins identified in a prior Olink MS study (4).
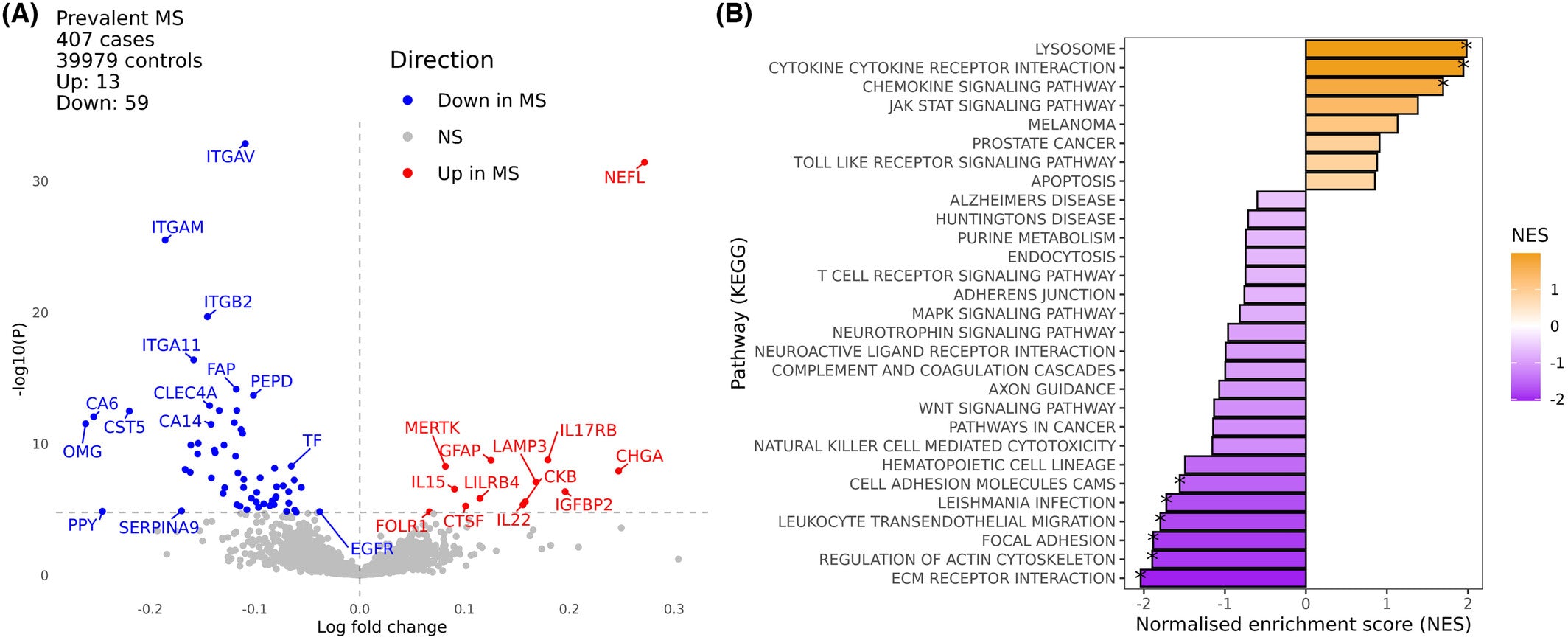
Figure. Plasma proteomic analysis of multiple sclerosis. (A) volcano plot of differences in plasma proteins in UK Biobank participants with (n = 407) and without (n = 39,979) MS at the time of sampling. (B) Gene set enrichment analysis of pathway level differences in the plasma proteome between MS and healthy controls. Source: Figure 1 from Jacobs et al. (3)
[The findings] demonstrate the power of biobank-scale datasets for discovering how the plasma proteome is altered in multiple sclerosis. Ultimately, this avenue of research could yield new drug targets, new insights into disease biology, and provide an adjunct to existing methods for individual-level prognosis in MS.
There is more to discover on neurological biomarkers. Click here to continue.
References
- Guo, Y., You, J., Zhang, Y. et al. Plasma proteomic profiles predict future dementia in healthy adults. Nat Aging 4, 247–260 (2024). https://doi.org/10.1038/s43587-023-00565-0
- Gan, YH., Ma, LZ., Zhang, Y. et al. Large-scale proteomic analyses of incident Parkinson’s disease reveal new pathophysiological insights and potential biomarkers. Nat Aging 5, 642–657 (2025). https://doi.org/10.1038/s43587-025-00818-0
- Jacobs, B.M., Vickaryous, N., Giovannoni, G., Proitsi, P., Waters, S. and Dobson, R. (2024), Plasma proteomic profiles of UK Biobank participants with multiple sclerosis. Ann Clin Transl Neurol, 11: 698-709. https://doi.org/10.1002/acn3.51990
- Åkesson, J., Hojjati, S., Hellberg, S. et al. Proteomics reveal biomarkers for diagnosis, disease activity and long-term disability outcomes in multiple sclerosis. Nat Commun 14, 6903 (2023). https://doi.org/10.1038/s41467-023-42682-9
Publications
Explore selected highlights from the referenced publications, including landmark neurology studies powered by UKB-PPP data.