Expanding access to high-quality NGS-based proteomics
Olink® Reveal now verified for sequencing on MGI T1+ and G400 sequencers
Olink® Reveal continues to expand access to high-quality, NGS-based proteomics. Verification studies have demonstrated that Olink® Reveal data can be sequenced on MGI T1+ and G400 platforms, using optimized protocols to deliver reliable and consistent performance.
Verification overview
The verification performed by Olink assessed the compatibility of MGI T1+ and G400 sequencers with Olink Reveal. The evaluation focused on the consistency of performance with Illumina’s NextSeq 2000 P4 flow cell, including read counts, quantitative correlations, assay variability, and detectability.
Results demonstrated that:
- Read yields on MGI sequencers were comparable to those obtained with the NextSeq 2000 P4 flow cell (Figure 1).
- High correlations indicated consistent quantitative performance between MGI and Illumina sequencing data (Figure 2).
- Coefficient of Variation (CV) differences between platforms were minimal, providing a solid basis to identify meaningful biological patterns (G400 mean CV relative to NextSeq 2000: +0.27%; T1+ mean CV relative to NextSeq 2000: +0.21%).
- Detectability, the proportion of proteins detected above limit of detection (LOD), was comparable across sequencers, indicating proteins are detected with similar reliability.
In addition, evaluation of two-library sequencing on the T1+ showed that two Olink® Reveal libraries can be processed simultaneously without significant impact on data quality or precision.
Collectively, these results demonstrated Olink Reveal has comparable quantitative performance, reproducibility, and detectability on MGI T1+ and G400 sequencers when compared with Illumina-based workflows.
By extending verified compatibility to MGI T1+ and G400 sequencers, Olink expands access to scalable, NGS-based proteomics—empowering more laboratories to generate actionable insights from 1,000 carefully curated proteins using their existing sequencing infrastructure.
Contact us to learn how to start running Olink® Reveal on your MGI T1+ or G400 sequencer.
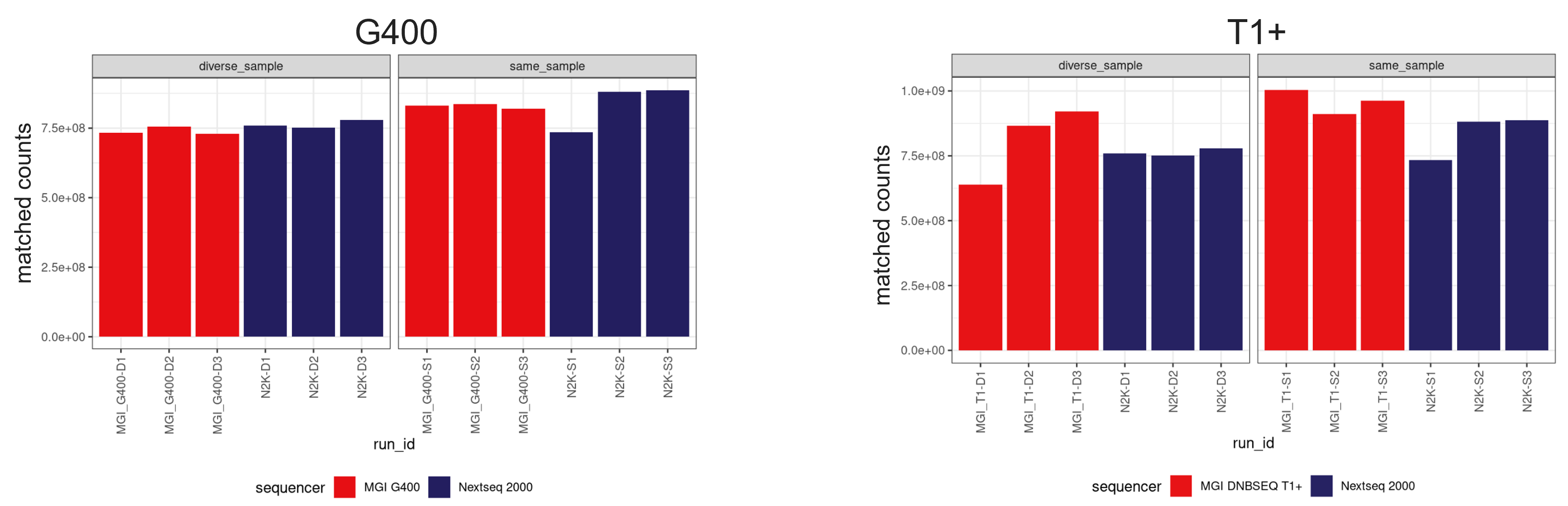
Figure 1: Bar charts comparing read yields across MGI T1+ FCL, MGI G400 FCL, and Illumina NextSeq 2000 P4 flow cells. MGI sequencers show comparable read counts to Illumina, confirming sufficient coverage for Olink® Reveal analysis.
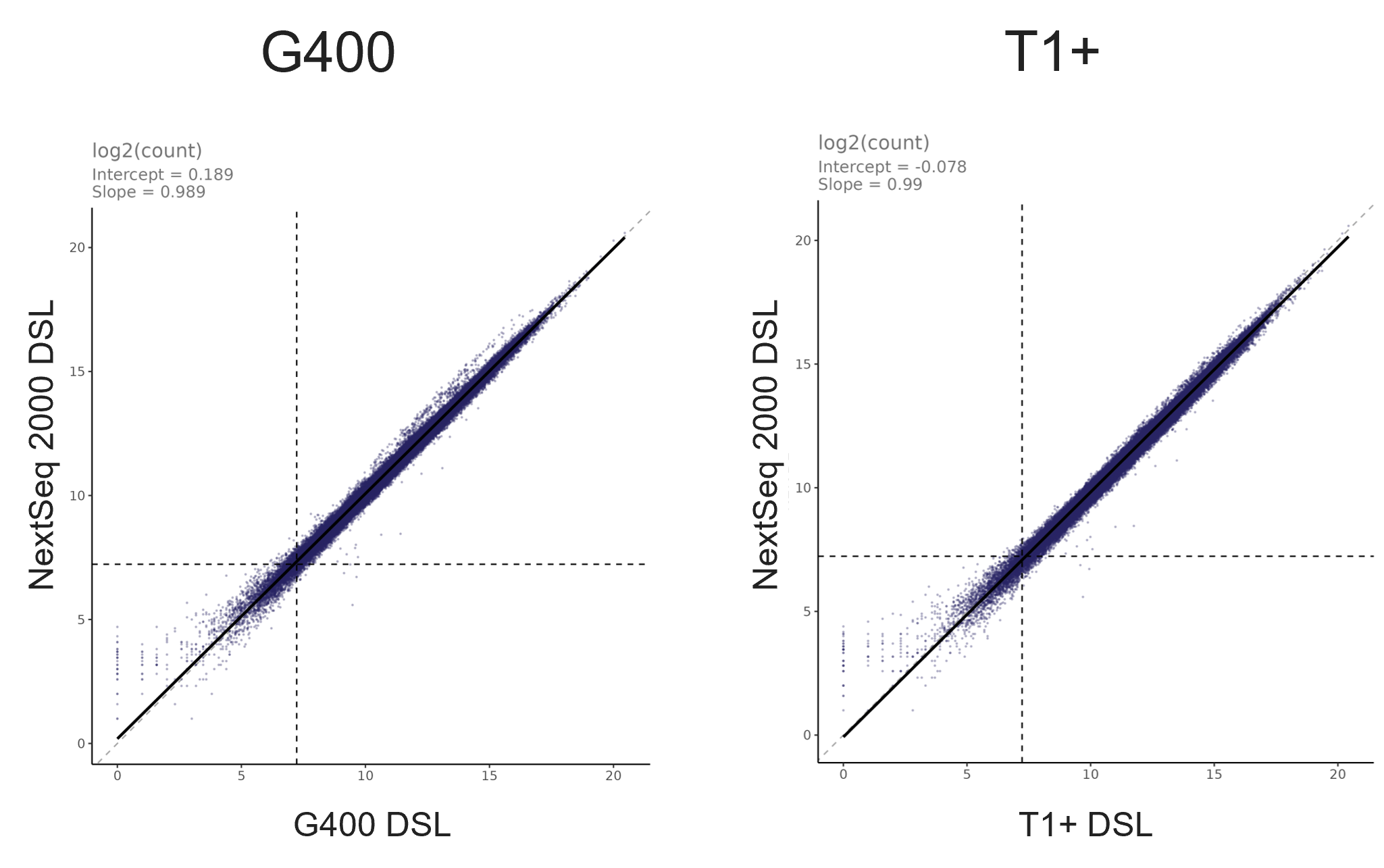
Figure 2: Scatterplots showing Pearson’s correlation between MGI (T1+ and G400) and Illumina NextSeq 2000 sequencing data for Olink® Reveal target proteins. High global correlation demonstrates quantitative equivalence between platforms, with no systematic differences observed.